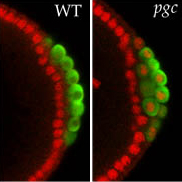
 Our long-standing interest in germ cell development began with identification of maternal effect genes that control the formation and specification of germ cell fate. Molecular characterization of these genes demonstrated that phylogenetically conserved, germ line-specific RNA regulators play an extensive regulatory role during all stages of the germ line life cycle. In particular, early on, we showed that localization of RNA and formation of RNA-protein particles at the posterior pole of the Drosophila oocyte and early embryo are critical for germ line specification and patterning of the embryonic axis. Today, as many as 250 different maternally-provided mRNAs have been observed to be enriched at the posterior pole. Proteins encoded by these “germ plasm effector” RNAs contribute to the formation, specification and differentiation of primordial germ cells. For nanos (nos) RNAs, we showed that only the localized pool is translated, while the unlocalized pool, which is evenly distributed throughout the egg and early embryo, is translationally repressed. More recently, we found that localization and translation of localized germ plasm RNAs is common and in general regulated by sequences in the 3’UTR. By employing single-molecule F
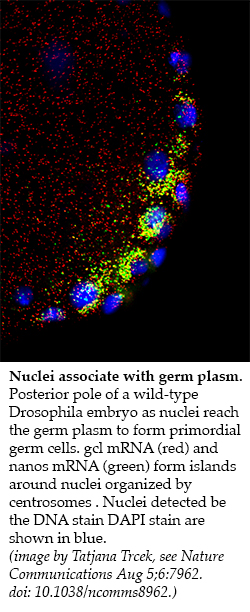
Our long-standing interest in germ cell development began with identification of maternal effect genes that control the formation and specification of germ cell fate. Molecular characterization of these genes demonstrated that phylogenetically conserved, germ line-specific RNA regulators play an extensive regulatory role during all stages of the germ line life cycle. In particular, early on, we showed that localization of RNA and formation of RNA-protein particles at the posterior pole of the Drosophila oocyte and early embryo are critical for germ line specification and patterning of the embryonic axis. Today, as many as 250 different maternally-provided mRNAs have been observed to be enriched at the posterior pole. Proteins encoded by these “germ plasm effector” RNAs contribute to the formation, specification and differentiation of primordial germ cells. For nanos (nos) RNAs, we showed that only the localized pool is translated, while the unlocalized pool, which is evenly distributed throughout the egg and early embryo, is translationally repressed. More recently, we found that localization and translation of localized germ plasm RNAs is common and in general regulated by sequences in the 3’UTR. By employing single-molecule F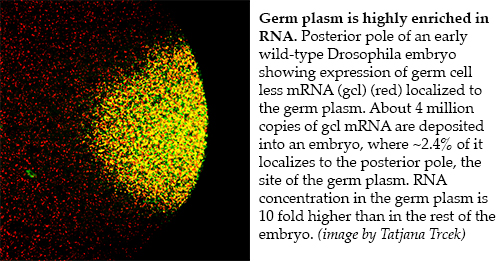 ISH, we have begun to analyze the spatial and temporal distribution of RNAs within the germ plasm. We find that the mRNA distribution within the germ plasm is structured and is organized with respect to the polar granules, conserved RNA-protein containing granules and hallmarks of germ cells. Some mRNA clusters are embedded deeply within a granule while others are more peripheral. Using biophysical, genetic and biochemical approaches as well as structure determination of individual protein components such as Oskar and Tudor in collaboration Rui-Ming Xu (Chinese Academy of Sciences, Beijing, China), we are interested in determining the mechanisms that dictates this distinct pattern of mRNA-protein organization.
ISH, we have begun to analyze the spatial and temporal distribution of RNAs within the germ plasm. We find that the mRNA distribution within the germ plasm is structured and is organized with respect to the polar granules, conserved RNA-protein containing granules and hallmarks of germ cells. Some mRNA clusters are embedded deeply within a granule while others are more peripheral. Using biophysical, genetic and biochemical approaches as well as structure determination of individual protein components such as Oskar and Tudor in collaboration Rui-Ming Xu (Chinese Academy of Sciences, Beijing, China), we are interested in determining the mechanisms that dictates this distinct pattern of mRNA-protein organization.
Research Areas:
|
|
|
|
|
|